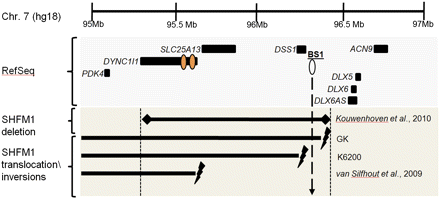
Chromosomal abnormalities at chromosome 7q21-23 associated with SHFM1. A schematic representation of the genomic positions of breakpoints from chromosomal rearrangements in individuals with SHFM1 mapped to human genome assembly 18 (hg18) and compared to the location of DYNC1I1 eExon 15 and 17. An 880-kb microdeletion in an individual with a split foot phenotype was found to be at 95.39–96.27 Mb (Kouwenhoven et al. 2010). In the GK family, the 46,XY,t(7;20)(q22;p13) translocation breakpoints mapped to chr 7: 96.2–96.47 Mb. In the K6200 family, the chromosomal inversion breakpoints mapped to chr 7: 96,219,611 and 109,486,136. The breakpoint coordinates of a 7:46, XY, inv(7) (p22q21.3) with SHFM1 and pervasive developmental disorder-not otherwise specified (PDD-NOS) was found to be at chr 7: 95.53–95.72 Mb (van Silfhout et al. 2009). All of these chromosomal abnormalities overlap with DYNC1I1 eExon 15 and 17 (orange ovals). Two of these chromosomal aberrations do not overlap with the BS1 AER enhancer (white oval). (Lightning bolts) Translocation and inversion breakpoints; (diamonds) deletion.











