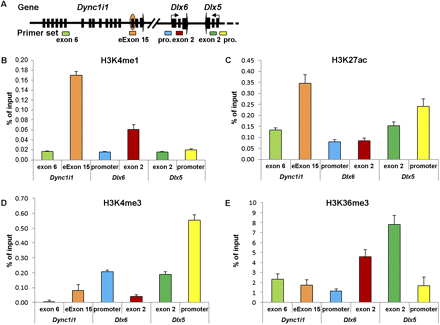
Histone modification signatures of the Dync1i1 eExon 15 in the mouse E11.5 limb bud. (A) Schematic representation of the Dync1i1-Dlx5/6 locus, showing the relative positions of primer sets used for ChIP-qPCR analyses: Dync1i1 exon 6 and eExon 15; Dlx6 promoter (pro.) and exon 2; Dlx5 exon 2 and promoter (pro.). (B) ChIP–qPCR analyses of H3K4me1, an enhancer histone mark. (C) ChIP-qPCR analyses of H3K27ac, an active enhancer histone mark. (D) ChIP-qPCR analyses of H3K4me3, a promoter histone mark. (E) ChIP-qPCR analyses of H3K36me3, a transcribed gene histone mark. (X-axis) Primer pairs; (y-axis) percentage of input recovery. (Error bars) SE from three technical replicates of a representative experiment.











