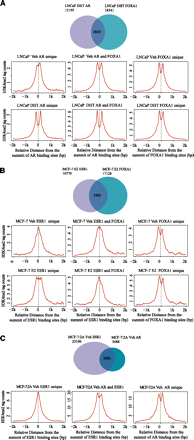
Mono-nucleosome level H3K4me2 ChIP-seq at nuclear receptor and FOXA1 binding loci in the MCF-7 (A), LNCaP (B), and MCF-7:2A (C) cell lines. (A) (Top panel) Venn diagram of AR binding in relation to FOXA1 binding. (Middle panel) Distribution of H3K4me2 signal centered on AR-unique, AR/FOXA1 shared, and FOXA1-unique sites in the unstimulated condition. (Bottom panel) Distribution of H3K4me2 signal centered on the AR-unique, AR/FOXA1 shared, and FOXA1-unique sites under conditions of androgen stimulation. (B) (Top panel) Venn diagram of ESR1 binding in relation to FOXA1 binding. (Middle panel) Distribution of H3K4me2 signal centered on ESR1-unique, ESR1/FOXA1 shared and FOXA1-unique sites in unstimulated cells. (Bottom panel) Distribution of H3K4me2 signal centered on ESR1-unique, ESR1/FOXA1 shared, and FOXA1-unique sites in estrogen stimulated cells. (C) (Top panel) Venn diagram of ESR1 binding in relation to AR binding. (Bottom panel) Distribution of H3K4me2 signal centered on ESR1-unique, ESR1/AR shared, and AR-unique sites in unstimulated cells.











