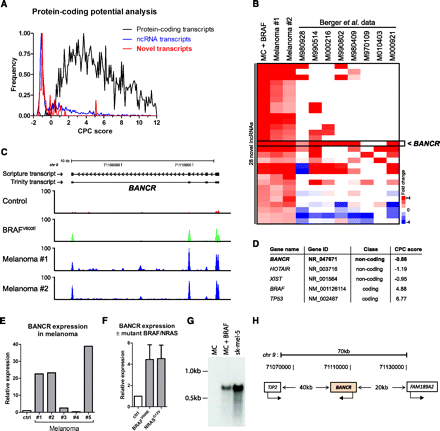
Oncogenic BRAFV600E regulates expression of many novel transcripts. (A) Frequency distribution plot showing coding potential analysis (CPC scores) of novel transcripts discovered by de novo assembly. One thousand five hundred random annotated ncRNAs and 1500 random protein-coding genes were also analyzed for reference. (B) Analysis of BRAF up-regulated novel lncRNAs in a publicly available melanoma RNA-seq data set. We remapped and reanalyzed Berger et al. (2010) data and derived expression values for novel transcripts up-regulated in all three of our samples. Robustly up-regulated novel transcripts (with negative CPC scores) were clustered with Berger et al. (2010) data revealing recurrently highly expressed lncRNAs. (C) Histograms of raw RNA-seq data in control sample, in melanocytes overexpressing BRAFV600E and in both primary melanomas (Mel. 1, Mel. 2). Scripture and Trinity assembly for BANCR is also shown. y-axis is number of RNA-seq reads normalized for mapping variation. (D) CPC score for BANCR. Examples of lncRNAs HOTAIR and XIST and protein-coding genes BRAF and TP53 are shown for reference. (E) BANCR expression in primary melanomas validated by qRT-PCR. (F) BANCR expression in melanocytes overexpressing BRAFV600E or NRASG12V measured by qRT-PCR. Data are means from three experiments ± SD. (G) BANCR Northern blot using RNA from control melanocytes (MCs), melanocytes overexpressing BRAFV600E, or sk-mel-5 melanoma cells. (H) Schematic of BANCR locus.











