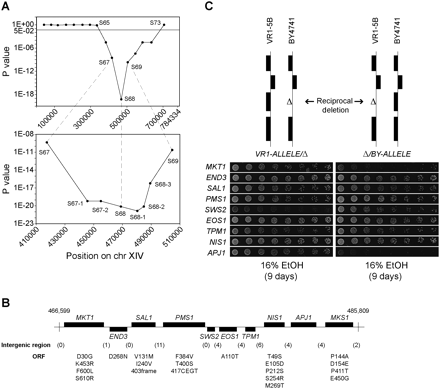
Fine-mapping and identification of the causative genes in QTL3. (A) The 87-kb locus defined by SNP markers S67, S68, and S69 in QTL3 showed the lowest probability of random segregation in 101 highly ethanol-tolerant segregants. Further fine-mapping was achieved by scoring five additional markers within the 87-kb interval in the same segregants. Calculation of the P-values revealed the strongest linkage for a 16-kb locus defined by markers S68, S68-1, and S68-2. (B) The name and location of each ORF in the fine-mapped locus is shown as annotated in SGD (Cherry et al. 1997). The interval from nucleotide 466,599 to 485,809 was sequenced in VR1-5B and BY4741, which revealed 115 polymorphisms, of which part were in intergenic regions (numbers in parentheses). For the ORFs, only polymorphisms that change the amino acid sequence are indicated (amino acid in BY4741, followed by position in the protein and amino acid in VR1-5B). SAL1 has a frame shift mutation in BY4741 resulting in an earlier stop codon and truncation of the protein, which is assumed to be a loss-of-function gene product (Dimitrov et al. 2009). PMS1 has an insertion of four amino acids at position 417 in VR1-5B. The sequence of BY4741 in this interval is the same as that of S288c (Cherry et al. 1997), except for one nucleotide in SAL1 that causes an amino acid change at position 131 (valine in BY4741 and methionine in S288c and VR1-5B). (C) Reciprocal hemizygosity analysis. For the nine genes in the fine-mapped locus, two diploid strains were constructed in the VR1-5B/BY4741 hybrid background that carried either the VR1-5B (left) or BY4741 (right) allele from the gene. The rest of the genome was identical between the two hybrids. The reciprocal deletions were engineered in the haploid strains, after which the proper haploids were crossed to obtain the diploid hybrids. The ethanol tolerance of the diploid hybrids was determined by scoring the growth of twofold dilutions on 16% ethanol after 9 d. This revealed different contributions of the parental alleles from MKT1, SWS2, and APJ1 to high ethanol tolerance. The strain pairs were always spotted on the same plate. The results were assembled from different plates, thus slight differences in growth may be present between hybrid pairs that otherwise do not show differences in ethanol tolerance. Hence, only growth differences between strains within a hybrid pair are relevant. The growth of the wild-type diploid hybrid was similar to that of the hybrid pairs whose ethanol tolerance was unaltered.











