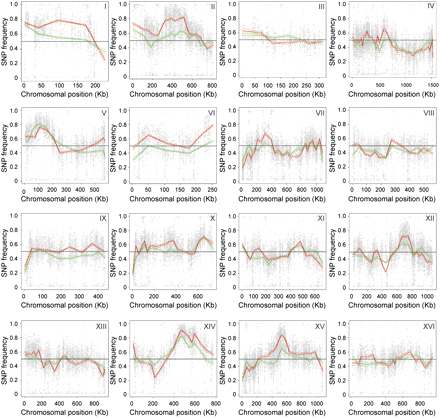
Genetic mapping of QTL involved in high ethanol tolerance by whole-genome sequence analysis. QTL were mapped by whole-genome sequence analysis of DNA extracted from a pool of 136 segregants tolerant to at least 16% ethanol (16% pool; green line) and from a pool of 31 segregants tolerant to at least 17% ethanol (17% pool; red line). The genomic DNA of the parents, VR1-5B and BY4741, and of the two pools, was sequenced and aligned to identify SNPs. The nucleotide frequency of quality-selected SNPs in the sequence of each pool was plotted against the chromosomal position. Significant deviations from the average of 0.5 indicate candidate QTL linked to high ethanol tolerance. Upward deviations indicate linkage to QTL in the ethanol-tolerant parent VR1-5B. The three major QTL on chromosomes V, X, and XIV are not significantly different between the two pools. However, in several instances, e.g., on chromosomes II, XII, and XV, minor loci can be identified, showing a significant difference between the two pools. These candidate QTL are more distinctive in the 17% pool compared to the 16% pool. The difference in SNP frequency between the two pools is certainly significant when the simultaneous confidence bands do not overlap.











