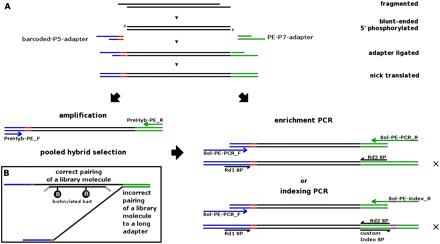
(A) Schematic overview of the library preparation procedure using the Illumina PE adapter (internal barcode in red). After a cascade of enzymatic reactions and cleanup steps, enrichment PCR can be performed to complete the adapter sites for Illumina PE sequencing (Rd1 SP, Rd2 SP are PE sequencing primers). Alternatively, libraries can be pooled for hybrid selection (if desired), and then enrichment PCR can be performed after hybrid selection. To achieve an even higher magnitude of pooling for sequencing, “indexing PCR” can be performed instead of “enrichment PCR,” whereby unique indices (in purple) are introduced to the adapter, and a custom index sequencing primer (index-PE-sequencing-Primer) is used to read out the index in a separate read. Finished libraries that have all the adapters necessary to allow sequencing are marked with an X. (B) Schematic figure of “daisy-chaining” during pooled solution hybrid capture, which may explain why a large proportion of molecules are empirically observed to be off-target when using long adapters. Library molecules exhibiting the target sequences hybridize to biotinylated baits, but unwanted library molecules can also hybridize to the universal adapter sites. The adapters of our “truncated” libraries (including barcode: 34 and 33 bp) are about half the length of regular “long” adapters (64 and 61 bases), and thus may be less prone to binding DNA fragments that do not belong to the target region.











