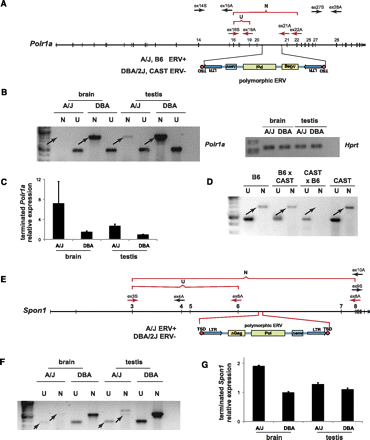
Disruption of additional genes by polymorphic, intronic ERVs in either orientation. (A) Genome structure of Polr1a containing a polymorphic AS ERV in intron 20, present in A/J and B6 and absent from DBA/2J and CAST mice. Various PCR primers are shown; (ex) exon number; (S) sense; (A) antisense. (Red arrows and brackets) cDNA amplicons; (U) upstream; (N) nonterminated, i.e., full-length. (B) Premature Polr1a termination occurs in brain and testis of A/J but not DBA mice. RT-PCR assays measured expression of upstream (U) versus nonterminated (N) transcripts, using ex16S and ex19A versus ex16S and ex22A primers, respectively. (Arrows) Differentially expressed, nonterminated transcripts. (Right) Loading control for spliced Hprt transcript assayed by RT-PCR. (C) Quantitative RT-PCR assay measuring relative differences between upstream and downstream Polr1a transcript levels, i.e., prematurely terminated transcripts. (Error bars) Range of duplicates. (D) Parent-of-origin effect on nonterminated Polr1a transcript levels in heterozygous mice. RT-PCR assays measured expression of upstream (U) vs. nonterminated (N) transcripts, using ex16S and ex19A versus ex16S and ex21A primers, respectively. (Arrows) Differentially expressed, nonterminated transcripts. See Supplemental Figure 6. (E) Genomic structure of Spon1 containing a polymorphic ERV in intron 6, present in A/J but not DBA mice. (F) Premature Spon1 termination in brain and testis of A/J but not DBA mice. (Arrows) Differentially expressed, upstream (U) and nonterminated (N) transcripts shown by RT-PCR assays. Both upstream and particularly full-length Spon1 transcripts are reduced in A/J mice (based on similar input RNA levels vs. DBA mice). (G) Quantitative RT-PCR assay measuring relative differences between upstream and downstream Spon1 transcript levels, i.e., prematurely terminated transcripts. (Error bars) Range of duplicates.











