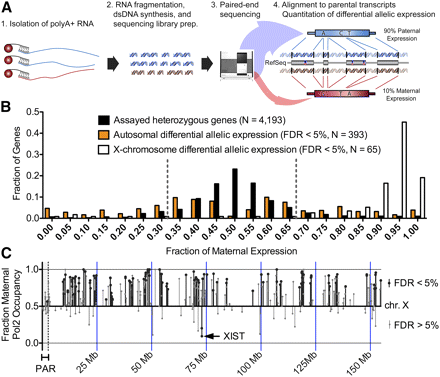
(A) Diagram of our method for using RNA-seq to measure differential allelic expression. First, poly(A)+ RNA was isolated using magnetic beads conjugated to oligo(dT) nucleotides. After RNA fragmentation, dsDNA was synthesized and subjected to paired-end sequencing on an Illumina Genome Analyzer. Reads were then aligned to GM12878-specific maternal and paternal versions of all RefSeq transcripts. Differential allelic expression was called when significantly more reads aligned to a single allele than would be expected by random. (B) Distribution of the fraction of maternal expression for all heterozygous genes (black), autosomal genes with differential allelic expression (orange), and X-chromosomal genes with differential allelic expression (white). (C) Prediction of differential allelic expression (y-axis) along the X chromosome (x-axis) using allelic occupancy of RNA Pol2. (Black lines) Significant differential allelic RNA Pol2 occupancy; (gray lines) nonsignificant binding. The shaded region on the left indicates the pseudoautosomal region that is not inactivated. All significant differential allelic occupancy predicted expression as expected. Genes that do not achieve statistical significance in the inactivated region of the X were a mix of genes that are known to escape inactivation as well as false negatives.











