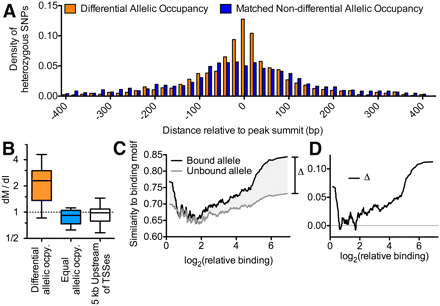
(A) Histogram of the distance of heterozygous SNPs from the location of maximal ChIP-seq signal for sites with (orange) and without (blue) differential allelic TF occupancy. To control for potential observation biases resulting from high read coverage at variants near the center of binding sites, the sites of equal allelic occupancy were chosen to match the differential allelic occupancy in two ways. First, for each site of differential allelic occupancy, we required the total number of aligned reads covering heterozygous variants in the matched site to be equivalent. Second, we required that the total number of variants in each binding site was also equivalent. If a suitably matched site did not exist, the site was excluded from the sites of differential allelic occupancy for this analysis. Using this strategy, the distribution of aligned reads at heterozygous variants was not significantly different between the sites of differential allelic occupancy and the matched set of equal allelic occupancy (P = 0.15, two-sided Wilcoxon rank-sum test). (B) The ratio of the rate of motif-disrupting to non-motif-disrupting intergenic mutations (dM/dI) across all sited of differential allelic TF occupancy (orange), and at TF binding sites that lack significant differential allelic occupancy (blue). To allow comparison with cis-regulatory DNA, the distribution of dM/dI is also shown for regions 5 kbp upstream of 10,000 randomly chosen TSSs (white). Whiskers show 95% confidence intervals. We excluded TFs for which we only observed a single motif-disrupting variant across all binding sites. (C) For the bound (black) or unbound (gray) allele at all sites of differential allelic occupancy, the similarity to TF binding motif (as a fraction of the optimal match) at sites of heterozygosity (y-axis) plotted against relative binding (the ratio of reads aligning to the bound vs. unbound allele; x-axis). Data were smoothed over a 32-data-point sliding window. The shaded region labeled Δ indicates the amount of difference in motif similarity between bound and unbound alleles, and is plotted in panel D.











