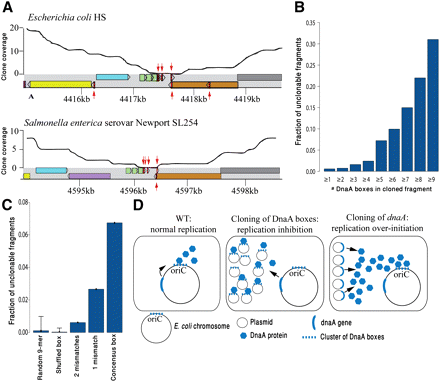
Toxic DNA binding sites. (A) Reproducibility of an intergenic gap in Enterobacteria. Coverage plots of a syntenic 5 kb in E. coli (refseq: NC_009800) and Salmonella enterica (refseq: NC_011080) are presented. Colored rectangles represent genes, with homologous genes sharing the same color. (Red arrows) DnaA boxes with up to one mismatch relative to the consensus sequence; (upper arrows) DnaA boxes on the forward strand; (lower arrows) DnaA boxes on the reverse strand. This region harbors the DnaA-titrating datA locus (Kitagawa et al. 1998). (B) Unclonability increases when multiple clustered DnaA boxes exist in a cloned DNA fragment. DnaA boxes distant up to 20 bp apart were clustered together. The horizontal axis denotes the minimal cluster size. (C) Unclonability increased with predicted affinity of the DnaA box to DnaA. Consensus DnaA boxes (TTATCCACA) are more unclonable than boxes with one or two mismatches. Data are shown for clusters containing two or more DnaA boxes. (D) A model for dnaA/DnaA-box toxicity. In normal conditions (left), DnaA is produced from a single locus and binds to the oriC to initiate replication. When DnaA box clusters are cloned on a high-copy plasmid (middle), they titrate cellular DnaA protein and inhibit replication. When additional copies of the dnaA gene are cloned (right), replication over-initiation occurs.











