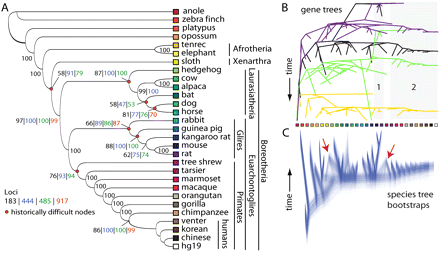
Evolutionary history of placental mammals resolved from conflicting gene histories. (A) Summary of STAR species trees generated from 183-locus and 917-locus data sets (Supplemental Table S1), in addition to the 444-locus data set that included UCEs from Stephen et al. (2008) and a 485-locus data set that included 41 exons (see Discussion). Note that STAR trees contain no branch length information. (B) Discord among four representative gene trees from the 183-locus data set. In general, gene trees were highly discordant, although some similarities emerged, such as the sister relationship between rat and mouse (shaded box 1) and monophyly of primates (shaded box 2). Discord among all gene trees is depicted in Supplemental Figure S7. (C) Widespread consensus among 1000 species-tree bootstrap replicates of the same 183-locus data set. STEAC trees (see Methods) are depicted because the branch lengths allow for better visualization of branching patterns, but STAR results supported the same topology. Cones emanating from terminal tips of species trees (red arrows) indicate disagreement among bootstrap replicates, for example, in the placement of the sloth and tree shrew. Colored squares indicate terminal taxa from A.











