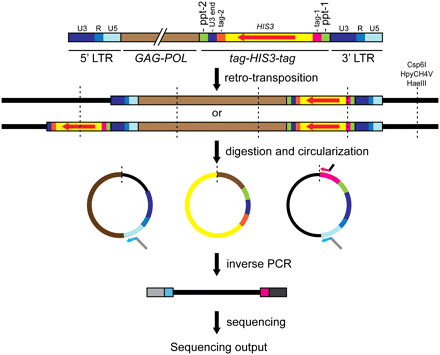
High-throughput sequencing of de novo Ty3 integration sites. Step 1: Expression of Ty3-ppt-HIS3 containing two unique 60-bp tags for amplification of integration joints. Step 2: Alternative reverse transcription products, dependent on position of ppt. Step 3: Restriction of genomic DNA with one of three restriction enzymes (Csp6I, HpyCH4V, and HaeIII) and circularization. DNA fragments flanking de novo Ty3 integration sites were selectively amplified by inverse PCR (iPCR) using primers that annealed to the Ty3 U5-end LTR and Ty3 tag-1. Primers contained Illumina adaptor sequences and the Ty3 LTR primer contained the Illumina sequencing primer sequence. Step 4: The iPCR products from separate reactions were combined and processed for sequencing using the Illumina GAIIx system. Ty3 transcription begins at the 5′ edge of R and continues through U5 into the internal domain and the 3′ U3 and R regions. Reverse transcription regenerates the complete U3-R-U5 LTR at each end of the cDNA. (Vertical dashed lines) Digestion/ligation sites; (red arrow) HIS3 ORF; (bent arrows) iPCR primers. Arrows indicate primers: (light blue) Ty3 U5-end LTR; (pink) tag-1; (gray) upstream Illumina adaptor with sequencing primer; (black) downstream Illumina adaptor.











