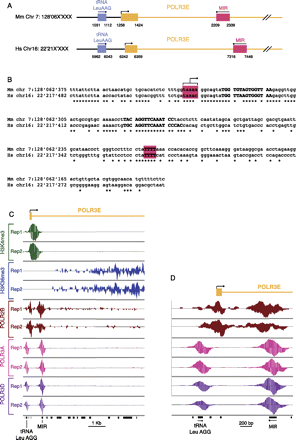
(A) Arrangement of RNAP-III genes in shared syntenic regions containing the Polr3e gene in the mouse and human genomes. The last four numbers of the chromosome coordinates are indicated below the lines. (B) Alignment of the mouse (Mus musculus, Mm) and human (Hs) MIR sequences. The region where RNAP-III transcription is likely to initiate and the terminator region are highlighted in red. Similarities to A- and B-box elements are indicated in bold. Identities are labeled with asterisks. (C) View of the tags obtained in the ChIPs performed with antibodies against the factors indicated on the left. The accumulations of + and − tags are shown above and below the lines, respectively. The location of the first exon and intron of the Polr3e gene is shown at top, that of the tRNA Leucine and the MIR at the bottom. (D) Zoom-in of the region around the Polr3e transcription start site.











