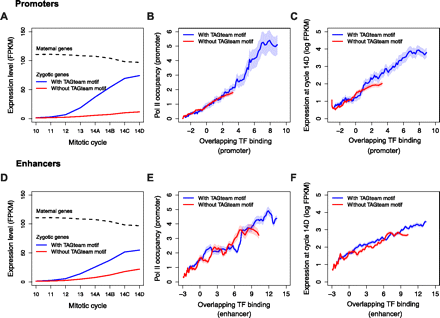
Binding of multiple factors in promoters and enhancers is correlated with Pol II occupancy of promoters and gene expression levels. (A) Zygotically transcribed genes with TAGteam motifs in their promoters (defined as the 1.5 kb centered on the TSS) have increasing expression from mitotic cycles 10–14. Maternally transcribed genes have slowly decreasing expression over the same time course (regardless of presence or absence of TAGteam motifs; data not shown). (FPKM) Fragments per kilobase per million mapped reads, a standard unit of gene expression measured by RNA-seq. (B) Pol II occupancy of promoters is correlated with binding in promoters. (C) Gene expression levels are correlated with binding in promoters. (D) Putative enhancers, defined as ChIP-bound regions 1.5–5 kb upstream of the TSS, with nearby TAGteam motifs (within 500 bp) are associated with increasing expression of downstream genes from cycles 10–14. (E) Pol II occupancy of promoters of downstream genes is correlated with binding in putative enhancers. (F) Gene expression levels are correlated with binding in upstream enhancers.











