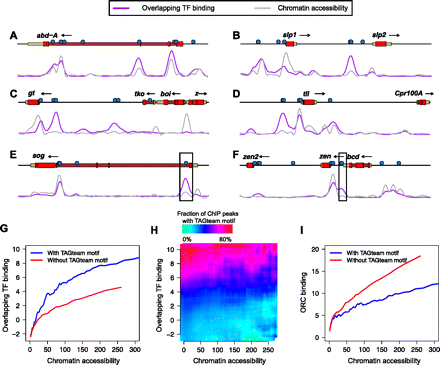
TAGteam motifs are associated with binding in different chromatin contexts. Levels of binding of all factors (purple) and DNase I digestion data (gray), both in arbitrary units, are plotted across 25 kb near the genes (A) abdominal A (abd-A), (B) sloppy paired 1 (slp1) and sloppy paired 2 (slp2), (C) giant (gt), (D) tailless (tll), (E) short gastrulation (sog), and (F) bicoid (bcd), zerknullt (zen), and zerknullt-related (zen2). Vertical axes differ between panels for clarity. Boxed TAGteam motifs have been previously demonstrated to be required for early expression (ten Bosch et al. 2006; Liberman and Stathopoulos 2009). ChIP and DNase I data were smoothed as described in Methods. (G) TAGteam motifs are associated with binding even after controlling for chromatin accessibility. Plot shows average overlapping binding at ChIP peaks with and without nearby (within 500 bp) TAGteam motifs after binning by DNase I signal. (H) TAGteam motifs are primarily associated with binding, not chromatin accessibility. Plot shows fraction of ChIP peaks with nearby TAGteam motifs, binned by both chromatin accessibility and levels of binding. (I) TAGteam motifs are depleted in regions with high ORC binding. Plot shows average ORC binding at ChIP peaks for the 21 transcription factors analyzed here with and without nearby (within 500 bp) TAGteam motifs after binning by DNase I signal.











