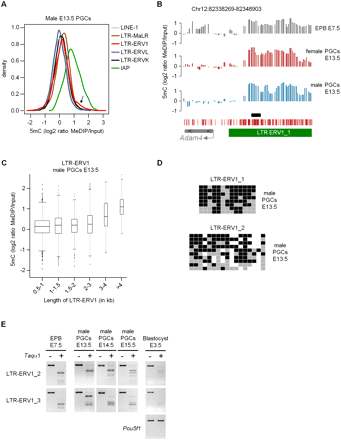
Cytosine methylation of repetitive elements in E13.5 PGCs. (A) The density plot represents the distribution of MeDIP log2 ratios of oligonucleotides located in various classes of repetitive elements in E13.5 male PGCs. (Arrow) Small proportion of probes in LTR-ERV1 elements with high MeDIP signals. The distribution is identical in female PGCs (Supplemental Fig. 11A). (B) Example of tiled LTR-ERV1 element on chromosome 12 that retains high levels of cytosine methylation in male and female E13.5 PGCs. The graphs show smoothed MeDIP over input ratios of individual oligonucleotides. (Black box) Position of the PCR fragment used for COBRA and bisulfite sequencing validations. The genomic position is indicated above the graphs. (C) The box plot shows the distribution of MeDIP log2 ratios in LTR-ERV1 elements in E13.5 male PGCs as a function of the size of LTR-ERV1 elements. Here and in subsequent box plots, the width of the box is proportional to the square root of the number of data points. The distribution is identical in female PGCs (Supplemental Fig. 11B). (D) Bisulfite sequencing confirms the presence of cytosine methylation in LTR-ERV1 elements in male E13.5 PGCs. In our conditions, the LTR-ERV1_1 primers amplify one single element, whereas the LTR-ERV1_2 primers amplify several elements with sequence polymorphisms. Squares represent CpG dinucleotides as unmethylated (gray), methylated (black), or absent (white). (E) COBRA shows that LTR-ERV1 elements are also methylated in PGCs isolated from male E14.5 and E15.5 embryos, as well as in preimplantation E3.5 blastocysts. Lack of methylation in the Pou5f1 (Oct4) promoter is used as a control for the purity of blastocysts. Note that the LTR-ERV1_3 primers, like the LTR-ERV1_2 primers, amplify several elements with minor sequence differences.











