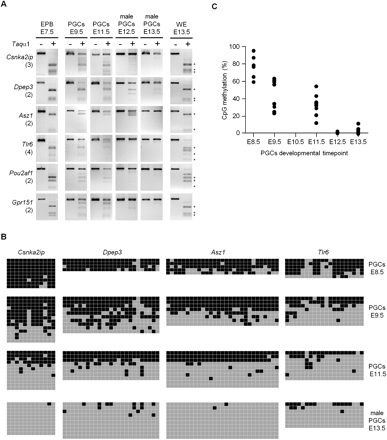
Kinetics of promoter DNA demethylation in developing PGCs. (A) DNA methylation of several gene promoters was analyzed by COBRA in E7.5 epiblasts (EPB) and PGCs isolated from E9.5 to E13.5 embryos. Methylation in E13.5 whole embryos (WEs) is shown as a control. Here and in other figures, the number of TaqαI sites in the amplified fragment is indicated in parenthesis, and asterisks mark restriction fragments representing end products of the digestion. (B) Bisulfite sequencing results in PGCs isolated from E8.5 to E13.5 embryos. Tested promoter sequences show high levels of cytosine methylation in E8.5 PGCs and progressive demethylation initiated in early PGC precursors. (Squares) CpG dinucleotides, either unmethylated (gray) or methylated (black). (C) Summary of the bisulfite sequencing methylation data from B and Supplemental Figure 8C. (Graph) Average percentage of cytosine methylation within the amplified fragments for every tested promoter.











