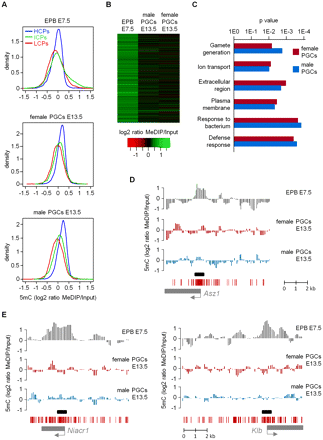
Erasure of promoter DNA methylation at all genes in E13.5 PGCs. (A) The density plots show the distribution of all gene promoters (classified as HCPs, ICPs, and LCPs) according to their average MeDIP ratios (calculated in regions covering −700 to +200 bp relative to the transcription start site). The small fraction of enriched promoters in E7.5 epiblasts (EPB) is absent in male and female E13.5 PGCs. (B) The heatmap shows that genes with a methylated promoter in E7.5 epiblasts lose the methylation signal in male and female E13.5 PGCs. (C) Ontology terms enriched in genes that lose promoter cytosine methylation in male and female E13.5 PGCs. (D) Demethylation of the promoter of the germline-specific gene Asz1 in E13.5 PGCs. (Graphs) Smoothed MeDIP over input ratios of individual oligonucleotides. (Black box) Position of the PCR fragment used for COBRA and bisulfite sequencing validations. (E) Demethylation of the promoters of somatic genes Niacr1 and Klb in E13.5 PGCs.











