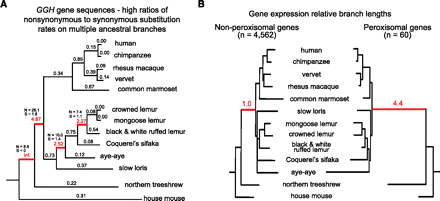
Positive and directional selection in the ancestral primate branch. (A) Ratios of the maximum likelihood-estimated (Yang 2007) rates of nonsynonymous (amino acid changing) to synonymous substitution (dN/dS) for the GGH gene shown directly above each branch. Values of dN/dS > 1, highlighted in red and with the number of estimated nonsynonymous (N) and synonymous (S) substitutions shown, are consistent with the past action of positive selection on several ancestral branches of the tree. (B) Relative gene expression branch lengths estimated from 4562 genes without peroxisomal functions and from 60 peroxisomal genes, considering genes with sufficient species representation for analysis of the ancestral primate branch (see Supplemental Methods). The ancestral primate branch, highlighted in red, is relatively 4.4 times longer among the peroxisomal gene set. Nine of the 33 top-ranked genes for patterns of expression consistent with directional selection on the ancestral primate lineage function play roles in the functioning of the peroxisome, significantly more than expected by chance (FDR = 7 × 10−9). The two phylogenies are plotted such that the sums of all branch lengths, excepting the ancestral primate lineage, are equal. The relative lengths of the ancestral primate branches of each phylogeny are shown (the value for the nonperoxisomal genes phylogeny was set to 1.0).











