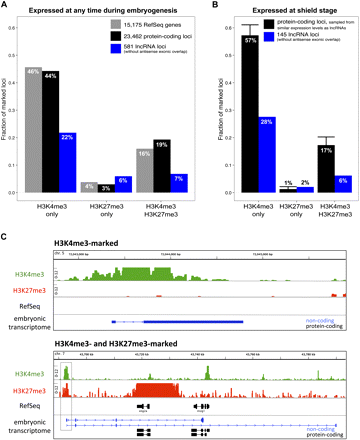
LncRNA genes carry chromatin marks associated with developmental regulators. Shown are the fractions of promoters (±500 bp relative to the transcription start site [TSS]) that are marked by a specific histone modification at shield stage. Histone marks were assessed by ChIP-seq experiments and analyzed for the presence of H3K4me3 only, H3K27me3 only, and both H3K4me3 and H3K27me3. RefSeq genes (gray bars); protein-coding loci (black bars); lncRNA loci (blue bars). (A) Marked fractions of promoters considering all loci. (B) Marked fractions of promoters only considering loci expressed at shield stage. In B, protein-coding loci were sampled from expression levels comparable to the set of 145 lncRNA loci expressed at shield (see Methods). Error bars, 1 SD of 10,000-times sampling. (C) Example chromatin profiles for a shield-expressed lincRNA gene marked by H3K4me3 (top) and for a lncRNA locus (overlapping the protein-coding genes eng2a and insig1) marked by both H3K4me3 and H3K27me3 (bottom). Signals are shown as the number of ChIP-seq reads that aligned overlapping in a 5-bp window (note that the y-axis ranges from 0–12).











