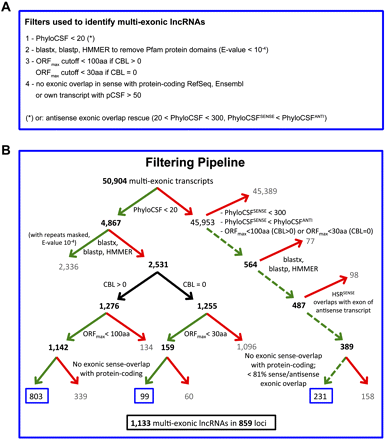
Overview of the stringent filtering pipeline that defined a conservative set of 1,133 lncRNAs. (A) Filters at a glance: overview of classification criteria used to define noncoding transcripts. (B) Detailed outline of the filtering pipeline that defined a conservative set of 1133 multi-exonic, embryonically expressed lncRNAs. The following filtering criteria were used: (1) Phylogenetic Codon Substitution Frequency (PhyloCSF) score <20 (left branch of the top node) or rescue by the antisense pipeline (right branch of the top node [dashed lines]: PhyloCSFsense < 300 and PhyloCSFsense < PhyloCSFanti and highest scoring region [HSR] overlapping with an exon on the opposite strand); (2) no known protein homologs based on blastx, blastp, and HMMER; (3) maximal ORF (ORFmax) <100 aa (transcripts with alignments [complete branch length (CBL) > 0]) or <30 aa (transcripts without alignments [CBL = 0]); and (4) no sense-overlap with any protein-coding transcript. At each step, a green arrow denotes the transcripts that passed the filter; a red arrow, those that were removed. Black bold numbers indicate the number of transcripts that passed the filter. Blue boxes highlight the number of transcripts that passed all filters and are considered noncoding (1133 lncRNAs in 859 loci).











