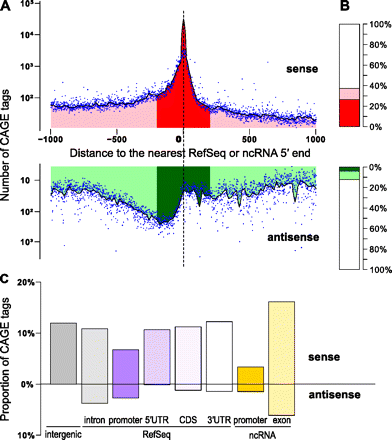
MOE transcription start sites recapitulate known transcript initiation and reveal the extent of noncoding transcripts. (A) The distribution of distances between nanoCAGE TSSs and the closest documented 5′ end of a transcript (RefSeq model or full-length noncoding FANTOM3 RNA). The number of CAGE tags is represented on different logarithmic scales for sense and antisense directions. The areas within 200 bp or 1000 bp are shaded in, respectively, darker or lighter shades of red (for TSS located on the same strand), or green (for TSS located on the opposite strand). (B) Proportion of TSSs located <200 bp (dark color), <1000 bp (light color), or >1000 bp (white) from RefSeq or FANTOM3 noncoding RNA transcripts, on the sense (red) or antisense (green) strand, both of which correspond to the distribution plotted in panel A. The white area completing the histograms depicts the remaining proportion of nanoCAGE TSS distant of >1000 bp from those documented transcripts' 5′ ends. (C) Histogram depicting the proportion of tags aligned to the proximal promoter of transcript models (defined as the region spanning from the 5′ end to 500 bp upstream), the 5′ UTR, the coding sequence (CDS), the 3′ UTR (in decreasing purple colors), the proximal promoter of FANTOM3 noncoding RNA (in orange), and the FANTOM3 noncoding RNA (in light orange). The upper part of the bar plot shows TSSs located on the same strand as the annotation, while the lower part depicts TSSs located on the opposite strand. (Gray bar) The percentage of TSSs that do not colocalize with any of those annotations.











