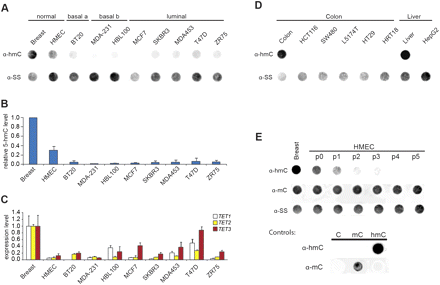
Reduced global 5hmC levels and TET1/2/3 gene expression in human cell lines. (A) Duplicate dot-blots of DNA (500 ng) from normal human breast tissue, a primary mammary epithelial cell line from non-cancerous tissue, and eight breast cancer cell lines probed with antibodies specific to 5hmC or ssDNA illustrate that global 5hmC levels are reduced in the DNA of primary and cancer cell lines relative to normal breast. (B) Inter-sample differences in global 5hmC levels by densitometric analysis of dot-blots shown in panel A. 5hmC values were normalized to the ssDNA loading control and scaled relative to normal breast DNA. Values are the means of two technical replicates. (C) RT-qPCR analysis of expression levels of TET1, TET2, and TET3 in normal human breast tissue, a primary mammary epithelial cell line, and eight breast cancer cell lines. Expression levels were normalized to GAPDH expression, and expression levels in normal human breast set to 1. Error bars represent the SD of two technical replicates. (D) Duplicate dot-blots of DNA (500 ng) from normal human colon and liver tissue and six cancer cell lines probed with antibodies specific to 5hmC or ssDNA illustrate that global 5hmC levels are also reduced in the DNA of colon and liver cancer cell lines relative to normal tissue. (E) Duplicate dot-blots of DNA (500 ng) from normal human breast tissue, and the primary mammary epithelial cell line derived from that tissue probed with antibodies specific to 5hmC, 5mC, or ssDNA show that global levels of 5hmC, but not 5mC, are gradually reduced upon transformation of normal breast tissue to cell culture. Ten nanograms of amplified mouse Tex19.1 promoter in which all cytosines were either unmodified (C), methylated (mC), or hydroxymethylated (hmC), was used as a control.











