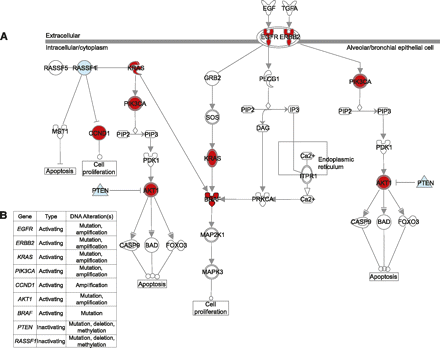
Activation of EGFR signaling in non-small-cell lung cancer (NSCLC) can occur via disruption of several different components at multiple levels of the pathway. (A) Different proteins in the EGFR pathway can be activated (red) or inactivated (blue) by underlying genetic or epigenetic changes at the DNA level, leading to aberrant pathway activity and oncogenic signaling in NSCLC. Examples of key oncogenes affected include EGFR, RAS, PIK3CA, and AKT. Conversely, examples of tumor suppressors that are inactivated include PTEN and RASSF1. (B) Genetic and epigenetic mechanisms responsible for the disruption of genes in the EGFR signaling pathway in NSCLC include DNA copy number alterations (amplification or deletion), point mutations, and DNA methylation changes. Thus, it is important to consider multiple aspects of the genome and epigenome simultaneously to elucidate the mechanisms driving pathway deregulation. This illustration was generated using Ingenuity Pathway Analysis software.











