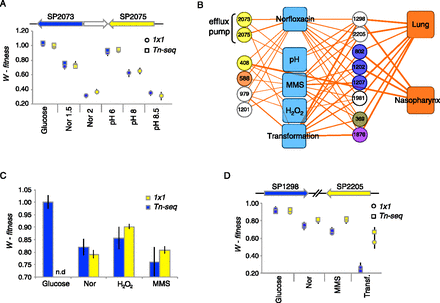
The identification of a dual-role efflux pump and the secondary DNA-damage response to Norfloxacin. (A) Tn-seq and 1 × 1 fitness data demonstrate that both genes in the efflux pump SP2073/SP2075 are involved in Norfloxacin resistance in a dose-dependent manner (concentrations 1.5 and 2 μg/mL are indicated by Nor 1.5 and Nor 2). Efflux pump mutants are also sensitive to pH stress, with a small but significant defect at pH 6 (P < 0.001) and an increasing defect at pHs greater than 8 (P < 0.001). Thus, the efflux pump appears to have a dual function—contributing to antibiotic resistance and to pH homeostasis. (B) A subnetwork of Norfloxacin responsive genes (color-coding same as in Fig. 2) indicates that 12 genes contribute to Norfloxacin resistance and also have interactions with the DNA-damaging stress conditions MMS and H2O2. Those genes that interact with MMS and H2O2 also often interact with transformation and have a defect in vivo in the lung and/or the nasopharynx. (C) Confirmation of the significant interactions of recN (SP1202) with Norfloxacin, H2O2, and MMS by 1 × 1 competitions (n.d., 1 × 1 fitness of the single deletion mutant in glucose was not determined). (D) Confirmation of the significant interactions of the DHH family genes SP1298 and SP2205 with Norfloxacin, MMS, and in DNA transformation by 1 × 1 competitions.











