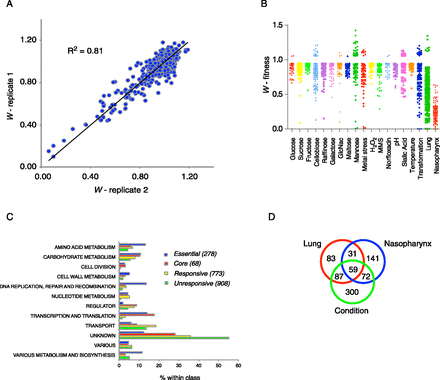
Condition-specific phenotypic profiling with Tn-seq generates a robust and novel data set. (A) Reproducibility of Tn-seq data was high (R2 = 0.63–0.85). Shown is a representative correlation between two libraries. (B) Significant average Tn-seq fitness scores for 17 in vitro conditions, lung and nasopharynx (also see Supplemental Table S1). (C) Classification of 2027 genes into four classes, which are divided into 12 functional categories. (D) Overlap of genes that respond to nasopharynx and/or lung and at least one in vitro condition.











