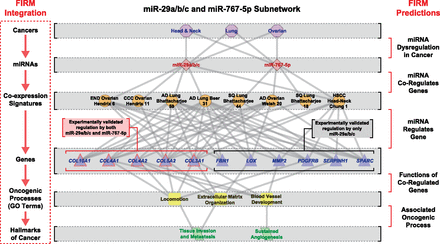
Summary of FIRM predictions for the miR-29a/b/c and miR-767-5p cancer–miRNA regulatory subnetwork. This subnetwork is included in both the metastatic- and cross-cancer–miRNA regulatory networks. The network is laid out hierarchically with (from the top down) cancers, miRNAs, coexpression signatures, genes that were experimentally validated through luciferase assays, significantly enriched GO biological process terms for the coexpression signature, and finally the GO terms associated with hallmarks of cancers. (Left) The FIRM integration strategy that is a flow of information through this hierarchy, where the red arrows indicate a FIRM prediction. The meanings of the FIRM predictions are described on the right side, where inference of a miRNA regulating a cancer coexpression signature predicts that the miRNA is dysregulated in that cancer. This same inference predicts that the miRNA regulates the genes in the signature, which can be tested experimentally. Functional enrichment of GO term annotations among the coregulated genes predicts the effect of regulating this set of genes, and association of the enriched GO terms with hallmarks of cancer predicts the oncogenic processes that might be affected.











