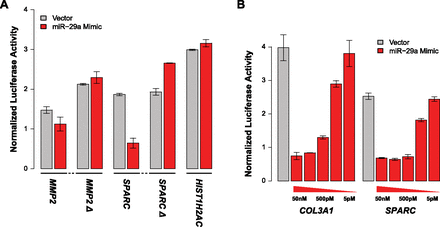
Luciferase reporter assay validation of miRNA-binding site predictions from FIRM. (A) Deletion of miR-29-binding sites ablates response to miR-29a mimic. The wild-type 3′ UTRs are MMP2 and SPARC. The miR-29-binding sites that deleted 3′ UTRs are MMP2 Δ and SPARC Δ. The deletions have a slight increase in normalized luminescence over their corresponding vector control, which is similar to what is observed for the negative control, HIST1H2AC, which does not have a miR-29-binding site. (B) Dose response curves for COL3A1 and SPARC titrating the amounts of miR-29a mimic (50 nM, 5 nM, 500 pM, 50 pM, and 5 pM).











