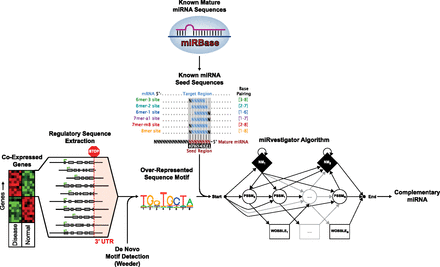
Overview of Weeder–miRvestigator tandem that we developed to identify miRNAs driving coexpression of transcripts. Quantitative assays of the transcriptome are used to identify gene-coexpression signatures comprised of genes with significantly similar gene-expression profiles. The 3′ UTR sequences for the coexpressed genes are then extracted from the genome and used as input into the Weeder algorithm. The Weeder algorithm searches the 3′ UTR sequences for an overrepresented motif, which is turned into a miRvestigator hidden Markov model (HMM). All of the miRNA seed sequences from the miRNA repository miRBase are compared with the HMM model of the overrepresented sequence motif using the Viterbi algorithm. The miRNA seed sequence with the most significant complementarity P-value is the most likely miRNA driving the coexpression signature and a hypothesis that can be tested experimentally.











