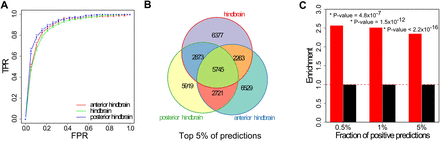
Hindbrain enhancers can be accurately predicted from DNA sequence. (A) Area under the ROC curve (AUC) for three Hb enhancer classifiers trained on three highly overlapping data sets (enhancers with activity in the anterior Hb, posterior Hb, and whole Hb). AUC values range from 0.5 (random discrimination) to a theoretical maximum of 1. We tested the performance of the classifiers in a cross-validation setting and obtained values of 0.89 (anterior Hb), 0.92 (posterior Hb), and 0.89 (combined Hb). (B) Overlap among the top-scoring 5% Hb enhancer predictions produced by all three Hb classifiers. (C) Fold-enrichment in 787 genes involved in Hb function in the neighborhood of positive predictions or putative Hb enhancers. Putative Hb enhancers were associated with the closest gene. P-values were computed using Fisher's exact test.











