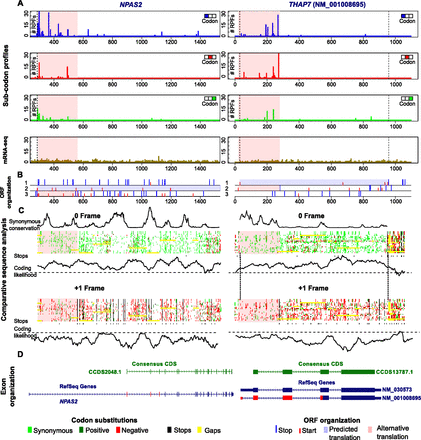
Dual coding in NPAS2 mRNA due to the presence of a translated nonupstream ORF and in THAP7 mRNA due to the overlap of the main ORF with an uORF. (A) Subcodon profile (top three rows) and mRNA-seq (fourth row) for NPAS2 mRNA (left; NM_002518) and THAP7 mRNA (right; NM_001008695). CDS coordinates are marked with dotted vertical lines. (B) ORF organization of NPAS2 mRNA (left) and THAP7 mRNA (right). The three reading frames are indicated as 1, 2, 3. Blue vertical lines indicate stop codons and start codons are indicated in red. Annotated CDS is shaded in light blue. The areas where translation in alternative frames is detected are shaded in light pink. (C) Comparative analysis of orthologous genomic sequences from 23 vertebrate species for NPAS2 (left) and from 19 vertebrate species for THAP7 (right). Colored bars represent codon substitutions within multiple sequence alignments for the standard (top) and alternative (bottom) reading frames (detailed alignments are in Supplemental Figs. S121, S122). Dark green and light green boxes correspond to synonymous and positive (in the BLOSUM62 matrix) substitutions, respectively; red boxes correspond to negative (in BLOSUM62 matrix) nonsynonymous substitutions. Gaps are shown in yellow and stop codons are in black. Stop codons are also aggregated across the entire alignment beneath each bar. Plots of coding likelihood are shown underneath the colored bars for both reading frames as calculated with MLOGD. Synonymous position conservation for the standard translation phase (pORF) is shown above the colored bar. (D) Exon organization of the NPAS2 locus (left) and the THAP7 locus (right). CCDS and RefSeq gene tracks from the UCSC Genome Browser are shown in green and blue bars, respectively. Alternatively decoded regions are indicated in red.











