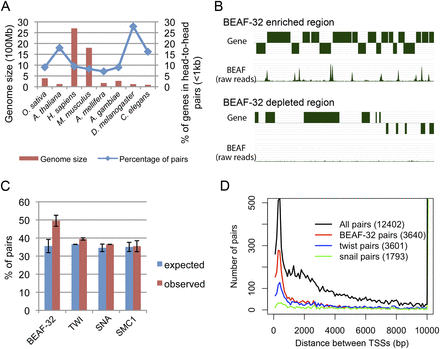
BEAF-32 specifically associates with close head-to-head gene pairs. (A) Genome size and percentage of genes in head-to-head gene pairs in different eukaryotic genomes. There is a high proportion of head-to-head gene pairs in the compact D. melanogaster genome compared with other species. (B) Snapshot of two regions of the D. melanogaster genome showing BEAF-32 binding sites associate with close head-to-head gene pairs. The top track represents genes. Genes above the line are transcribed from the plus strand and genes below the line are transcribed from the minus strand. The bottom track represents sites of BEAF-32 localization in the region; signal corresponds to the number of raw reads from ChIP-seq analysis. (C) Percentage of head-to-head gene pairs flanking different proteins. BEAF-32 associated pairs are significantly enriched for head-to-head gene pairs compared with the genome-wide expectation as well as compared with other proteins. The error bars are from the results of different data sets. The expected and observed fraction of gene pairs was calculated independently for each data set, and the mean and standard deviation were then determined. For BEAF-32, we used data sets obtained using embryos from this study and modEncode, and S2 cells. For TWI or SNA, we used data sets from different biological repeats. For SMC1, we used data sets for cell lines Kc, S2, and Bg3. (D) Distribution of distances between TSS's for genes flanking BEAF-32 and transcription factors. The number in parentheses is the total number of gene pairs in each category. BEAF-32 frequently associates with adjacent gene pairs close to each other.











