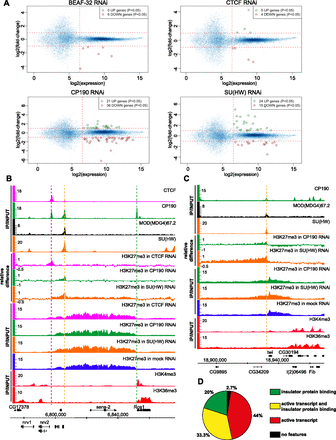
Functional effects of insulator protein withdrawal. (A) Affymetrix GeneChip expression analysis of cells from the RNAi experiments described in Figure 3. The average fold change between the two specific and two mock RNAi experiments (y-axes) was plotted against the highest average expression value detected in the mock or specific RNAi samples (x-axes). Each graph point represents one transcript interrogated by the microarray. Transcripts robustly expressed before or after specific RNAi treatment are to the right of the vertical dashed lines. Of these, those showing consistent twofold or greater change after specific RNAi treatment in both replicate experiments are circled. (B) As evident from ChIP-chip of H3K27me3 from mock RNAi-treated BG3 cells, the sens-2 gene is repressed by PcG. The right border of the corresponding H3K27me3 domain is sharp and coincides with a standalone CP190 site (marked by a vertical green dashed line) and with the Rca1 transcript. The ChIP-chip with H3K4me3 and H3K36me3 indicates that Rca1 is transcriptionally active. The left side of the H3K27me3 domain declines gradually with no obvious border. It harbors gypsy-like and CTCF+CP190 binding sites marked by orange and purple dashed lines, respectively. The knock-downs of insulator proteins have no effect on the position of the right border of the H3K27me3 domain but change the shape of its left tail. The changes in histone methylation profile are best seen on the relative difference browser tracks. (C) twi is also repressed by PcG mechanisms in BG3 cells. The right border of the corresponding H3K27me3 domain is set by the presence of an active transcript. The left border is maintained by a gypsy-like (class 3) insulator (vertical orange dashed line), as evident from the extension of K27 trimethylation after SU(HW) or CP190 knock-down. (D) The pie chart shows the frequencies of various genomic features associated with definable H3K27me3 domain borders.











