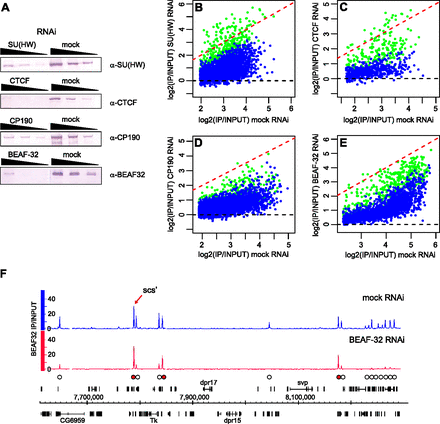
The effects of RNAi knock-down on the binding of insulator proteins to chromatin. BG3 cells were subjected to RNAi against key insulator proteins followed by ChIP-chip. (A) Western blots of threefold serial dilutions of nuclear protein from cells treated with specific and mock dsRNA (indicated above the panels) show 10-fold or greater knock-down of the corresponding proteins. The antibodies used for detection are indicated to the right, and the loading controls are shown in Supplemental Figure S4. The comparison of average binding for (B) SU(HW), (C) CTCF, (D) CP190, and (E) BEAF-32 after mock and specific RNAi shows that the binding is reduced at the majority of sites (data points below red dashed line). (Blue dots) The sites with consistent reduction in both replicate experiments (estimated conservatively with unpaired t-test; z-scores < −3); (green dots) others. (F) scs′ is one of the BEAF-32 high-affinity binding sites resistant to RNAi. The BEAF-32 ChIP-chip signals after BEAF-32 and mock RNAi are plotted along the segment of chromosome 3R. (White circles) Peaks affected by BEAF-32 knock-down; (red circles) peaks that remain unchanged. The genes shown above the coordinate scale are transcribed from left to right, those below the scale from right to left.











