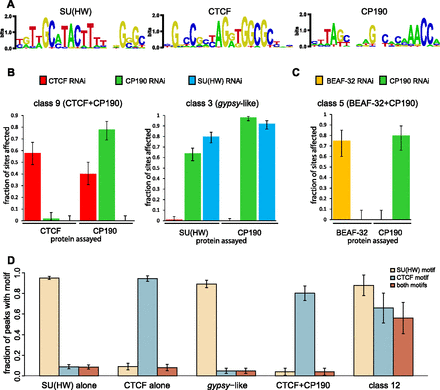
The sequence determinants and interdependence of the insulator protein binding to chromatin. (A) The logo representations of sequence motifs characteristic of SU(HW), CTCF, and CP190 binding sites defined by the MEME algorithm and used in the analysis in D. (B) The effects of the RNAi knock-down on the target protein and its co-binding partners. The sites at which ChIP-chip signal was consistently reduced judged from the comparison of two replicate mock RNAi experiments and two specific RNAi experiments (z-scores < −3, unpaired t-test) were counted and their fractions plotted. Here and in C and D, the error bars indicate the 95% confidence interval. The bar plots show that the binding of CP190 to some of the class 9 but not at gypsy-like sites depends on CTCF. However, the binding of CTCF to class 9 sites does not depend on CP190. In contrast, the binding of SU(HW) and CP190 to gypsy-like sites is interdependent. (C) As illustrated by this bar plot, BEAF-32 and CP190 bind to common sites independently. (D) The presence of SU(HW) and CTCF recognition sequences within indicated classes of sites demonstrates that the coincidence of the two motifs is responsible for the co-binding of SU(HW) and CTCF to class 12 sites.











