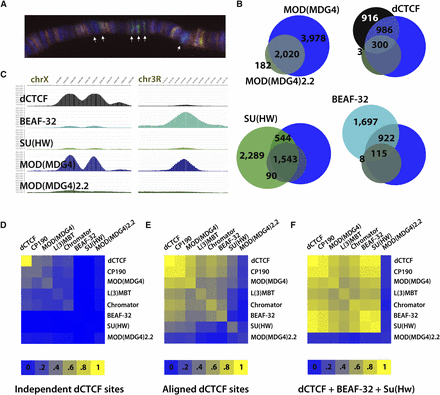
dCTCF and BEAF-32 recruit isoform(s) of MOD(MDG4) different from MOD(MDG4)2.2. Aligned dCTCF sites are enriched for MOD(MDG4) and additional cofactors. (A) Immunofluorescence microscopy of MOD(MDG4) (green) and MOD(MDG4)2.2 (red) on Drosophila polytene chromosomes. MOD(MDG4) staining includes many discrete bands not accounted for by MOD(MDG4)2.2 specific antibodies, depicted with white arrows. (B) Genome-wide overlap of dCTCF, BEAF-32, and SU(HW) with MOD(MDG4) and MOD(MDG4)2.2. Many dCTCF (45%) and BEAF-32 (34%) sites overlap MOD(MDG4) isoform(s) not represented by MOD(MDG4)2.2. Meanwhile, many SU(HW) (37%) sites overlap MOD(MDG4)2.2 sites, as expected. (C) ChIP-seq profile for MOD(MDG4) and MOD(MDG4)2.2 reveals many unique peaks specifically in the MOD(MDG4) profile, accounted for at BEAF-32 and dCTCF sites. (D–F) Heatmap representation of percentages of dCTCF sites in which CP190, MOD(MDG4), MOD(MDG4)2.2, BEAF-32, SU(HW), L(3)MBT, and/or Chromator co-occur at independent dCTCF sites, aligned dCTCF sites, and sites where dCTCF aligns with both BEAF-32 and SU(HW).











