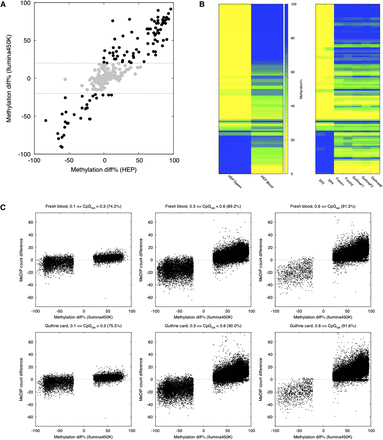
Array and sequencing-based DNA methylomics of Guthrie cards. (A) Comparison of Illumina450K-based “fresh cord blood vs. sperm” tDMRs with “blood vs. sperm” tDMRs identified in the Human Epigenome Project (HEP) (Eckhardt et al. 2006). Plotted are 468 genomic regions (∼200–300 bp in length) common between the HEP and the Illumina450K data sets. Illumina450K-based “fresh cord blood vs. sperm” tDMRs that display <20% methylation differences are shown in gray. R2 = 0.90 (overall), R2 = 0.50 (<20% methylation), both Pearson's. (B) A comparison of fresh cord blood and Guthrie card Illumina450K profiles with HEP data. Shown is methylation data for regions called “blood vs. sperm” tDMRs in the HEP. Each column represents profiles from a separate individual, and 106 different tDMRs are shown. (SP) Sperm. (C) Normalized MeDIP-seq read counts for the “blood vs. sperm” tDMRs identified by the use of Illumina450K-based fresh blood (top) or Guthrie cards (bottom) across a range of CpG densities. Only >20% tDMR methylation differences are shown. The percentage agreement is indicated in each panel. Number of different CpGs represented in each panel: Low CpG, n = 6404; mid-CpG, n = 28,384; high CpG, n = 18,135. Note the numbers are the same for the “Fresh blood” and “Guthrie card” data sets as the differentially methylated CpG sites were called on the Illumina450K arrays.











