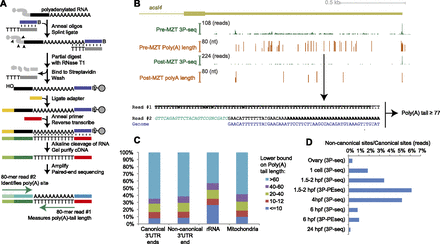
Genome-wide estimation of poly(A)-tail lengths. (A) Outline of 3P-PEseq. See text for description. (B) Poly(A) sites and poly(A)-tail lengths pre-MZT and post-MZT at the acsl4 3′ UTR. The 3′ UTR shown is as annotated in Ensembl v66. The height of the 3P-Seq plots shows the number of 3P tags at each position, normalized to the maximum value, which is indicated at the top of each axis. The height of the 3P-PEseq plots shows the average poly(A)-tail length measured at each position. The length is zero at positions for which no 3P-PEseq tags were obtained. (Arrow) The poly(A) site corresponding to the paired reads shown below. (Bold) Untemplated nucleotides. In this example, the cleavage position is within three genomically encoded A's, and thus, at least 77 of the 80 T's of read #1 correspond to untemplated A's of the the poly(A) tail. The adapter sequence in read #2 is in green italics. (C) Distribution of pre-MZT poly(A)-tail lengths at poly(A) sites mapping to the indicated loci and RNA classes. Canonical 3′ UTR ends are tallied as 3P-PE tags that map within 20 nt of a 3′ UTR end annotated using samples from other stages. Noncanonical 3′ UTR ends are tallied as 3P-PE tags that map within a 3′ UTR annotated in other stages but not within 20 nt of a poly(A) site defined at any other stage. rRNA and mitochondrial ends are tallied as tags that map within rRNA repeats and the mitochondrial chromosome, respectively. (D) Abundance of noncanonical isoforms. Plotted is the ratio of 3P or 3P-PE tags mapping to noncanonical 3′ UTR ends (defined as in C) relative to those mapping to canonical 3′ UTR ends.











