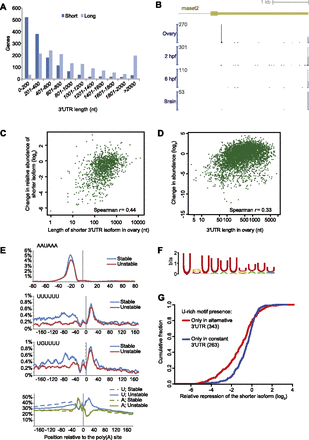
Differential accumulation of transcripts with short 3′ UTRs in the ovary and pre-MZT embryo. (A) Comparison of 3′ UTR lengths of the short and the long isoforms in genes with exactly two isoforms in the ovary. (B) Poly(A) sites of rnaset2 in the indicated samples. The shown 3′ UTR structure is as annotated in Ensembl v66. The height of each plot indicates the number of 3P tags ending at each position, normalized to the maximum value, which is indicated at the top of each axis. (C) Relationship between length of the shorter isoform and relative abundance of the shorter isoform in the pre-MZT embryo, as inferred from 3P tags for genes with two alternative poly(A) sites in the ovary. (D) Relationship between the length of the 3′ UTR in the ovary and the change in mRNA observed in the pre-MZT embryo relative to that in the ovary, as inferred by 3P tags. Analysis was for genes with a single 3′ UTR supported by at least 20 3P tags in the ovary. (E) Frequency of the indicated motifs or nucleotides flanking poly(A) sites of isoforms that were not reduced in the pre-MZT embryo compared to the ovary (stable) and those that were reduced at least twofold (unstable). For the hexamer motifs, the frequencies shown at each position are averages of a window of 11 consecutive nucleotides centered at that position. (F) The motif identified by Amadeus (Linhart et al. 2008) as significantly enriched in regions upstream of the stable sites. (G) Destabilization in the pre-MZT embryo of shorter isoforms lacking the U-rich motif. Genes with two UTR isoforms in the ovary and a U-rich motif 20–90 nt upstream of only one of the poly(A) sites were stratified based on the location of the motif—upstream of the distal site (red line) or upstream of the proximal site (blue line). A U-rich motif was defined as present if a decamer with up to two mismatches from the UK(U)8 consensus appeared 20–90 nt upstream of the poly(A) site.











