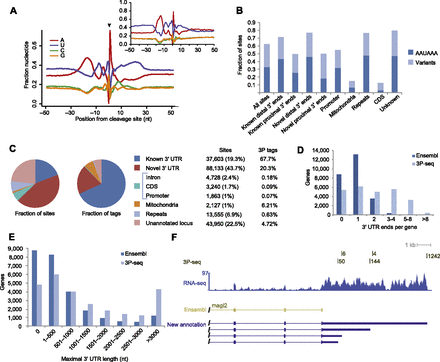
Reannotation of zebrafish 3′ UTRs. (A) Nucleotide sequence composition around all 197,350 3P-seq–identified poly(A) sites. (Black arrow) Cleavage position. As previously noted (Jan et al. 2011), the sharp adenosine peak at position +1, the depletion of A at position –1, and blurring of sequence composition at other positions was partly due to cases of cleavage after an A, for which the templated A was assigned to the poly(A) tail, resulting in a –1-nt offset from the cleavage-site register. (Inset) Sequence composition around poly(A) sites in C. elegans (Jan et al. 2011), redrawn for comparison. (B) Frequencies of sites containing the canonical PAS motif AAUAAA or one of its ten common variants in the region from –40 to –10 relative to the poly(A) site. Known distal 3′ ends are the distal-most poly(A) sites annotated in Ensembl, and known proximal 3′ ends are all other annotated poly(A) sites. Novel distal 3′ ends are poly(A) sites more distal than the distal-most annotated 3′ end. All other novel 3′ ends were designated as proximal. (C) Classification of poly(A) sites as fractions of sites or as fractions of the 3P tags. The poly(A) site classification scheme is described in Supplemental Figure S1. (D) Genes with alternative 3′ UTR isoforms in Ensembl v66 and following 3P-seq-based annotation. (E) Maximal 3′ UTR lengths in Ensembl v66 and following the 3P-seq-based annotations. For the new models, the longest 3′ UTR was supported by at least 10% of the 3P tags in at least one sample. (F) 3′ UTRs annotated for the magi2 gene. 3P-seq and RNA-seq tracks indicate all tags mapping to this locus. No 3′ UTR was annotated for this gene in Ensembl v66.











