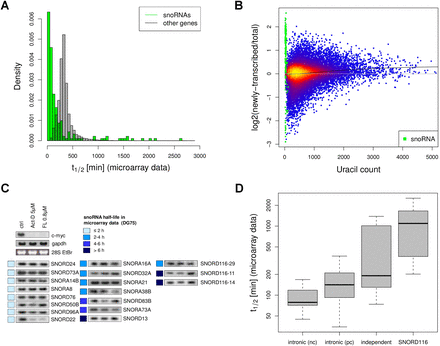
Poor processing efficiency of many, but not all, human snoRNAs. (A) Distribution of RNA half-lives (t1/2) based on the DG75 microarray data for snoRNAs (green) compared with all other genes (dashed). (B) Comparison of the transcript's uracil count against the log2 (newly transcribed RNA/total RNA ratios) allows the identification of reduced capture rates for short, i.e., uracil-poor, transcripts. The colors indicate the local density of points (yellow, high density; blue, low density). For genes with several transcripts, uracil number was calculated as the average of all transcripts. If no bias in capture rate were observed, log ratios should average around zero (gray line), indicative of no association of RNA half-life with uracil content. Local regression (black line) suggests a slight bias in this example, reflecting reduced capture efficiency of small transcripts, as seen in previous studies. For snoRNAs (green), log ratios are considerably higher than expected for their short length. (C) Northern blot data for 15 snoRNAs after transcriptional inhibition by treatment with either 5 μM Act-D or 800 nM FL for 6 h. 28S EtBr served as loading control. For almost all snoRNAs, expression levels after transcriptional inhibition remained unaltered, indicative of their high stability. (D) Distribution of RNA half-lives based on the DG75 microarray data for snoRNAs with different expression strategies: intronic (nc), snoRNA expressed as part of an intron of a noncoding gene; intronic (pc), snoRNA expressed as a part of an intron of a protein-coding gene; independent, snoRNA with independent promoter; SNORD116, SNORD116 cluster snoRNAs.











