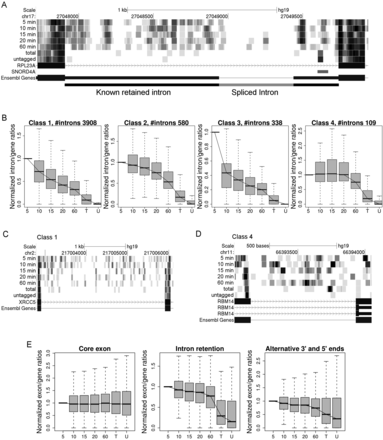
Characterization of distinct intron splicing kinetics. (A) Decay of an alternative splicing isoform of the RPL23A gene. Read densities for 5- to 60-min 4sU-RNA as well as total and untagged RNA are shown in shades of gray. This figure and all others showing read densities were created using the UCSC Genome Browser (Kent et al. 2002). The corresponding UCSC Genome Browser session containing read density values for all 525 genes analyzed can be accessed via http://www.bio.ifi.lmu.de/en/4sU-seq. Exons are indicated by boxes, introns by lines. Regions that are part of noncoding transcripts are indicated by boxes of smaller height. The known retained intron not translated is additionally indicated in the last line in black. The intron marked in gray is absent in all 4sU-RNA samples (RPKM < 0.5), which is indicative of very fast splicing. In total and untagged RNA, the retained intron is also absent, indicating either nonsense-mediated decay of the alternative splicing product or a secondary splicing event. (B) Higher level clustering of the intron clusters identified in the first clustering step (see also Supplemental Fig. 2). Clustering was performed based on intron/gene ratios of cluster representatives normalized to 5-min 4sU-RNA levels. Four classes of intron processing were identified: (Class 1) “Normal” decay with a linear slope in the first 20 min; (Class 2) reduced decay rates; (Class 3) increased decay rates; (Class 4) retained introns that are stable during the first 60 min but are eventually spliced and/or degraded. (C) Read density in all samples for an exemplary Class 1 intron. (D) Read density for an exemplary Class 4 intron. (E) Exon/gene ratios in all samples for core exons, exons constituting retained introns, as well as alternative 3′ and 5′ ends. Again, exon/gene ratios were normalized to ratios in 5-min 4sU-RNA.











