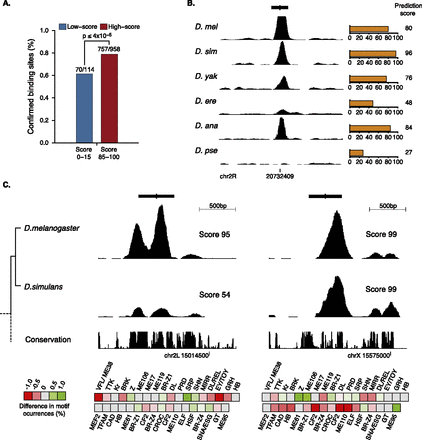
Predicting Twist binding across Drosophila species. (A) Sequences orthologous to D. melanogaster early Twist binding sites in five Drosophila species are significantly more often bound by Twist if they had high prediction scores (red; low-scoring sites are blue). For each of the 407 early Twist binding sites in D. melanogaster found by two independent ChIP studies (Zinzen et al. 2009; He et al. 2011), we classified the orthologous sequences from Drosophila simulans, Drosophila erecta, Drosophila yakuba, Drosophila ananassae, or Drosophila pseudoobscura with a SVM trained in D. melanogaster only (for details, see Methods). We then assessed whether the sites were bound in vivo using data from He et al. (2011) (note that the majority of D. melanogaster binding sites are bound across species as reported by He et al. 2011, leading to a high overall binding rate). Shown is the fraction of bound sites for the best and worst scoring sites (score ranges: 0–15 versus 85–100). (B) Prediction scores (orange) and ChIP-seq signals (normalized read density; black density track) correlate well across six different Drosophila species for a Twist binding site (black bar) close to the genes Dll and CG3650. (C) Examples of loss (left) or conservation (right) of a Twist binding site between D. melanogaster and D. simulans that had been correctly predicted despite largely similar (left) and different (right) overall motif content, respectively. The motif content is shown as a heatmap in which gray represents motifs with identical counts in both species (14 for the left vs. 10 for the right example, respectively) and shades of red and green represent smaller or higher motif counts in D. simulans, respectively. The UCSC phastCons track indicates sequence conservation across 14 insect species (Siepel et al. 2005). Consistent with the motif content heatmaps, the binding site sequence on the left is overall more highly conserved across species: 38.3% (left) versus 8.7% (right) of all nucleotides have a perfect phastCons score of 1.0, and the overall nucleotide identity between D. melanogaster and D. simulans was 86.2% (left) and 84.8% (right), respectively.











