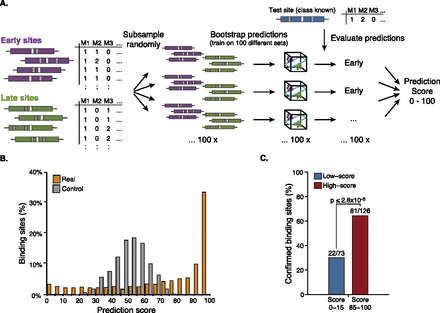
A prediction score for individual TF binding sites. (A) Schematic overview of the bootstrapping approach used to calculate a prediction score for each individual binding site. We excluded a site for testing (blue) and subsampled the remaining sites (left) 100 times to obtain 100 different training sets (middle), each of which we used to predict the test site (right) (see Fig. 2 for details). The prediction score represents the number of correct predictions and ranges from 0 to 100. (B) Distribution of the prediction scores for the classification of early versus late Twist binding sites (orange) and for controls for which we repeated the entire procedure after randomizing the sites' assignments to the early and late classes (gray). (C) Predictability of binding might be underestimated because sites with low prediction scores are less often reproducible by ChIP: Twist binding sites with high prediction scores (red; score-range 85–100) are significantly more often detected in an independent ChIP-seq data set (from He et al. 2011) than sites with low scores (blue; score range 0–15). Shown is the number of total and confirmed binding sites for sites in both score ranges and the binomial P-value.











