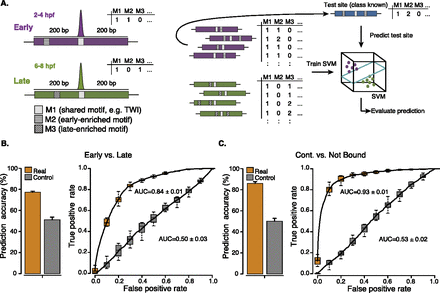
Cis-regulatory motif content predicts TF binding. (A) Schematic overview of the approach. To computationally classify context-specific TF binding sites based on their motif content, we counted all instances of known cis-regulatory motifs (denoted here as M1, M2, M3, etc.) in 401-nt-long windows centered on each ChIP-chip peak's summit (left; purple and green denote early and late bound sites, respectively). We used the motif counts (M1, M2, M3, etc.) for all binding sites as feature vectors for binary classification with a support vector machine (SVM) using leave-one-out cross-validation (LOOCV). Briefly, we excluded each binding site in turn for testing (blue), trained the SVM on the remaining sites, and predicted the test site (right). (B,C) Prediction (binary classification) of early versus late Twist binding sites (B) or continuously bound versus not bound Twist motifs (C) based solely on the motif content. (Orange) Prediction accuracies (percent of correctly classified sites; left) and receiver-operating-characteristic (ROC) curves (right; area under the curve [AUC] values are indicated). We repeated the entire procedure after randomizing the sites' class assignments (controls; gray), which yields random classifications (∼50%; AUC∼0.5) as expected (see Supplemental Discussion). The accuracy and AUC values for all other mesodermal TFs can be found in Supplemental Table S3; and for TFs, cofactors, and histone modifications in C. elegans, mouse, and human in Table 1.











