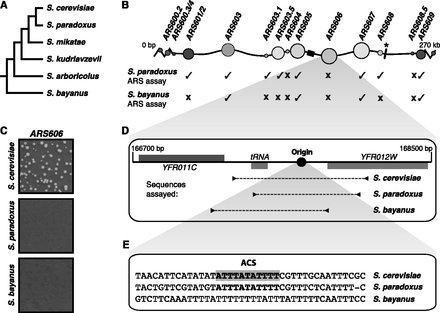
Functional conservation of replication origins on chromosome 6. (A) Phylogenetic relationship between sensu stricto yeast species (not to scale). (B) Schematic of S. cerevisiae chromosome 6 illustrating the location of replication origins (origins represented by circles; circle area corresponds to origin efficiency in W303; later origin activation time is represented by darker shading) (Yamashita et al. 1997), the centromere (black box), and a translocation relative to S. bayanus (black bar marked *). Locations equivalent to S. cerevisiae replication origins were assayed for ARS activity in S. paradoxus and S. bayanus. Ticks indicate that the replication origin is functionally conserved in the related species; crosses, that it is not. (C) Example assay plates for S. cerevisiae ARS606 and equivalent locations from S. paradoxus and S. bayanus. (D) Schematic of the gene structure around ARS606 and the fragments assayed for ARS activity in the indicated species. (E) Alignment of the ARS consensus sequence (ACS) from the indicated species.











