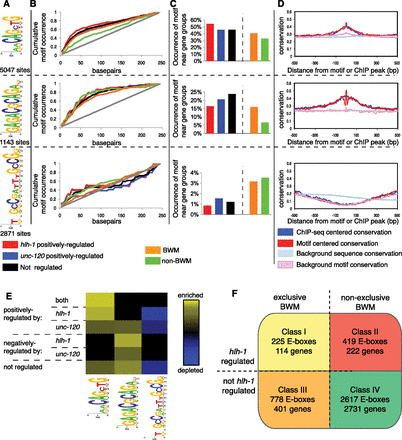
HLH-1-associated motifs correlate with directional expression control. (A) The web logo position-specific frequency matrix (PSFM) diagrams for three representative motifs and the accompanying number of sites identified near HLH-1 occupancy (250-bp radius). (B) The relative locations of three motifs compared to their experimentally identified binding sites (analyzed per Ozdemir et al. 2011). The AACAGCTG motif is centered on the called ChIP-seq peak (50% within ±25 bp of the peak for hlh-1 positively regulated or muscle-enriched genes). The GAGACGCAGA motif (second panel) is less central (within 75 bp). The GCCGatttGCCG motif (third panel) shows no significant centrality. The gray line represents a uniform distribution. (C) The occurrence of each motif within ±250 bp of the HLH-1 occupancy peak near genes (ChIP regions within 5 kb of a gene TSS) belonging to expression groups is shown. The E-box shows the greatest enrichment for genes characterized as hlh-1 positively regulated (first panel). The GAGACGCAGA motif is more closely associated with unc-120 positively regulated genes (second panel), whereas the GCCGatttGCCG motif is enriched near genes absent in BWM (third panel). (D) The conservation across sequenced nematodes (elegans, briggsae, remanei, and brenneri) of ChIP-seq identified regions with the three motifs is shown. Conservation around the in vivo binding (blue) and around the motif (red) is shown compared to background (light blue and pink) (Ozdemir et al. 2011), with higher values representing a higher level of conservation. The E-box and GAGACGCAGA motifs, along with their surrounding sequences, are strongly conserved, while the GCCGatttGCCG motif is not at all conserved. (E) Heat maps show the level of motif enrichment (yellow) or depletion (blue) for the CAgCTGtt, GAGACGCAGA, and GCCGatttGCCG motifs near broadly expressed genes that are similarly regulated (y-axis). The E-box is enriched near genes positively regulated by hlh-1 and unc-120. The GAGACGCAGA motif is enriched near genes negatively regulated by hlh-1 and positively regulated by unc-120. The GCCGatttGCCG motif is depleted near genes positively regulated by either factor. (F) There are four classes of E-boxes observed: Class I contains muscle E-boxes that are bound by HLH-1, and it is predicted that mutation of these sites will lead to changes in expression, as the nearby genes are both specific to BWM and regulated (positively or negatively, in contrast to the Archetypal Genes, which are exclusively positively regulated) by hlh-1; Class II contains E-boxes that are similarly functional but are near genes not exclusively expressed in BWM; Class III contains E-boxes that are not required for expression but likely make contributions to nearby genes that are expressed exclusively in BWM; and Class IV contains seemingly nonfunctional E-boxes that are not required for expression or associated with BWM expression.











