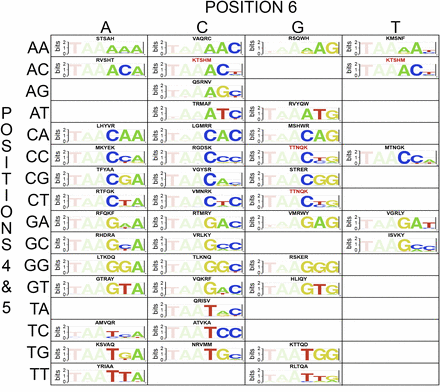
Selected HDs with favorable recognition preferences for each target site. A grid illustrating the selected HD variants that preferentially recognize or are compatible with particular 3′ binding site sequences. The amino acids that are present at the randomized recognition positions (43, 46, 47, 50, and 54) are indicated above each motif. Sequences in red indicate those that are present in more than one grid position (i.e., are compatible with two different sites). Empty boxes indicate the absence of quality HD recognizing these sequences.











