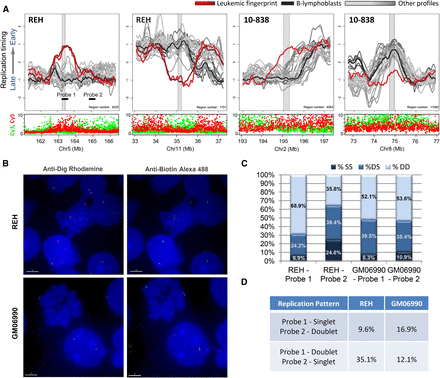
Replication-timing differences in karyotypically normal regions. (A) Shown are sample-specific fingerprint regions in cell line REH and patient 10-838 that lack genetic lesions under karyotypic analysis (both samples, with 10-838 being karyotypically normal), total Cy3 + Cy5 intensity (both samples) or Sanger CGH (REH), and therefore represent apparently epigenetic timing changes. Such regions may be explained by changes in long-range interactions or by subkaryotypic or CGH-resolution rearrangements. As in Figure 4, fingerprint profiles (REH or 10-838) are highlighted (red) against a background of other leukemic samples (gray) and B-lymphoblastoid cells (black). (B) Fluorescent in situ hybridization images of cell lines REH and GM06990 showing region-specific binding in metaphase nuclei and doublet/singlet hybridization patterns in interphase nuclei. (C) Quantification of observed singlet/singlet (SS), doublet/singlet (DS), and doublet/doublet (DD) configuration of allelic homologs for each probe shown in A. Only nuclei displaying at least one doublet allele (189 GM06990 and 296 REH) in either probe were scored, which may exaggerate the percentage of nuclei that appear to have replicated the regions asynchronously (single-doublets). (D) Quantification of the frequency with which one probe appeared to replicate prior to the other as a percentage of total chromosomes scored for which cis-linked probes 1 and 2 show a singlet–doublet configuration (378 GM06990 and 592 REH). In REH, probe 1 appears to replicate prior to probe 2 nearly 75% of the time, whereas in GM06990 either probe may replicate first.











